File Size: 61 MB
infiniSee is your Chemical Space navigation platform. Based on similarity, infiniSee finds molecules of interest in screening libraries or Chemical Spaces of infinite size. Given a template or query molecule, infiniSee returns unexpectedly similar molecules.
Let’s start with a thought experiment: If you fill an Olympic-size swimming pool with sand, you need a volume equal to approximately 10,000,000 cups of coffee. This volume, in turn, contains approximately 1013 grains of sand. Now, think of these individual grains of sand to represent possible drug candidates and you are looking for the one golden molecule. High-throughput virtual screening (HTVS) usually involves around five to ten millions of compounds which would be the same as looking for one specific grain in five to ten cups of sand. But who can guarantee that the perfect molecule is in one of the 5-10 cups you take from the pool? Chances at best are 1 in a million… Wouldn’t it be better and more promising if you can search in the whole pool or even thousands of them?
Discover unseen possibilities
Find molecules in unprecedented, large spaces of 1014 structures and more, or search your own in-house library for actives.
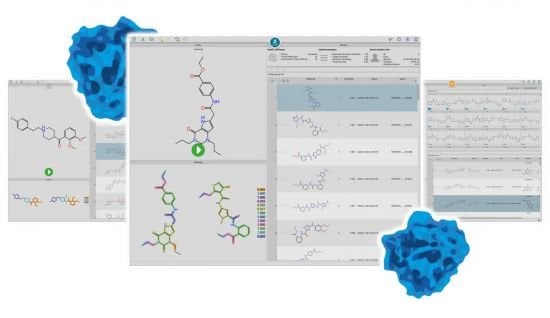
similarities infinisee
Similarity between a query and hit molecules is pleasingly visualized for you to select compounds effortlessly. infiniSee will show you why it considers something alike.
neighbors infinisee Distant neighbors in action
Mine compound libraries for interesting scaffold hops. infiniSee finds molecules which are distant at first sight, yet very close in chemistry and pharmacophore-based action.
Unlimited accessibles
From virtual to vial within days! Only what is highly likely to be formed in the lab will be proposed during infiniSee’s Chemical Space navigation. Collaboration with our partners allows delivery of desired hits within few weeks.
Find hidden similarities
With infiniSee you can finally compare apples and oranges. The underlying concept of infiniSee is strikingly easy: Instead of searching already “assembled” molecules, we instead perform a combinatorial build-up of compounds from “fragments”. Sources of fragments can be either combinatorial libraries or any fragment-generating procedure. infiniSee opens the possibility to screen billions of compounds through its similarity search technology by navigating vast Chemical Spaces and searching for distant neighbors of a query molecule. Results will be delivered within typically less than a minute on a standard laptop. Optionally, you can define fuzzy pharmacophores to increase the diversity of scaffolds or request important molecular structures to be present in the hits.
Your molecule can be anywhere
The ingenious twist of infiniSee is the possibility of searching not only in colossal Chemical Spaces, but the ability to also comb through spaces of different origins. The number of identical structures in sets from different spaces was found to be extremely low (Lessel et al. 2019) due to the design and setup of the spaces, as well as the diversity of the employed building blocks. No matter how big your in-house library and no matter how many compounds you acquire to add to it, it will only be a tiny fraction of what your chemists are capable of synthesizing. Involving distinct Chemical Spaces increases your chances of finding accessible molecules you would have missed otherwise.
fast blue
Navigate through vast Chemical Spaces at unprecedented speed.
visual blue
Understand similarity in a glimpse, with intuitive color-coding.
easy blue
Self-explanatory interface. Simply drag your query and get started.
Homepage
Download rapidgator
https://rg.to/file/39f2aa6bbf7367dccc6f3dcf881b744f/BioSolvetIT.infiniSee.v6.2.0.rar.html
Download nitroflare
https://nitroflare.com/view/DBC005FEA89D890/BioSolvetIT.infiniSee.v6.2.0.rar
Download 百度网盘
链接: https://pan.baidu.com/s/1Ysj4jIqU1G_nTDRlZKnbIg?pwd=svba 提取码: svba